Download PDF
How mountains of data—and growing momentum toward data sharing—are beginning to transform the research landscape.
Ophthalmology has a strong legacy of designing high-quality, randomized controlled trials (RCTs) and of driving clinical innovation. Now, throughout medicine, new approaches to research are emerging—from the launch of registries that will yield big data to the recently announced preliminary guidelines for data sharing from the International Committee of Medical Journal Editors (ICMJE). What are the implications for how research will be conducted and funded?
Randomized Controlled Trials: Creating Data
The first true RCT in ophthalmology, said David W. Parke II, MD, Academy CEO, was conducted from 1951 to 1953 by ophthalmologist Arnall Patz, MD, and pediatrician Leroy E. Hoeck, MD. It examined the link between oxygen levels and blindness from retinopathy of prematurity (ROP) in premature infants.

BEYOND RCTS. Dr. Patz funded his trial with a loan from his brother-in-law. The prices of today’s RCTs are driving innovators to explore other research strategies.
Early successes. “Some people think of this as the first and most important randomized trial in medicine,” said Dr. Parke. “It subsequently led to a huge change in the way premature infants were treated in neonatal ICUs.”
Another important trial was the Diabetic Retinopathy Study (DRS), which took place in the 1970s and tested whether laser treatment was effective in retarding the progression of blindness from diabetic eye disease. The results shook preconceived ideas held by some of the greatest luminaries in the field, said Dr. Parke. “DRS showed that high-quality randomized clinical trials can prove [that] our own inherent biases are wrong, and it provided data to make a meaningful, relevant, and clinically impactful decision. It opened the floodgates to a whole host of RCTs in ophthalmology that greatly improved patient care.”
An investment. Although we continue to gain a wealth of information from RCTs, they require substantial investments in time and expense, said Emily Y. Chew, MD, deputy director of the division of epidemiology and clinical applications at the National Eye Institute (NEI), which widely shares its publicly funded information. For example, she said, “Most diseases in ophthalmology, such as macular degeneration or diabetic retinopathy, are chronic, which means we need to follow these patients over time. In our studies, we have less than 3% loss of follow-up, which is great, but slow progression means studies require much time and money.” In addition, she said, sufficient funding is needed to make sure trials are well executed with adequate sample sizes.
Challenging to conduct. RCTs are also cumbersome, said Dr. Parke. “Enrolling patients can sometimes be difficult, and the very nature of preparing a trial and going through the various stages can occasionally mean that trial results are not available until after science has moved on.”
Further questions. Each trial also identifies new questions, Dr. Parke said. “You may start out with a trial on diabetic retinopathy, but you end up with new questions: What about macular edema? What about proliferative disease? What about when both exist together? Do we now have the money to fund 3 new trials?”
Applicable in real life? Perhaps the greatest limitation of RCTs is making the trial match what happens in daily life, said Wiley A. Chambers, MD, deputy division director at the Food and Drug Administration (FDA). “People will sometimes act differently when they know they’re in a trial and are being monitored,” he said. “This affects how generalizable the results will be.”
Dr. Parke added that the results achieved in RCTs are applicable in the real world only if the physician faithfully follows the trial protocol, which doesn’t always happen in the clinic.
 |
|
VARIATION. It may be challenging for clinicians to get results similar to those achieved under the strict protocols of a randomized controlled trial.
|
Meta-analyses: Combining Data
By combining data from several published trials, researchers conducting meta-analyses can derive new insights or validate and strengthen earlier findings, said Anthony P. Adamis, MD, senior vice president of ophthalmology at Genentech.
Advantages. “They can potentially provide greater [statistical] power to detect signals than is possible with an individual trial,” he said, giving the example of meta-analyses of anti-VEGF trials that evaluated the drugs’ systemic safety.
Dr. Parke added that if a meta-analysis is done in a biostatistically careful fashion, it may produce a more useful assessment of clinical reality than can a single clinical trial. However, Dr. Chew noted, the strength of the findings from a meta-analysis depend on the quality of the original data.
Drawbacks. Sometimes trials are inappropriately combined, said Dr. Chambers. “If results are sufficiently disparate—once data are combined—you have averages that look different from the original trials.” Dr. Adamis added, “The eligibility criteria and methods could have varied across the trials,” in which case it’s inappropriate to combine the data. Further, he noted that because negative results don’t always get published, there is a potential for selection bias, making the meta-analysis less than representative.
 |
|
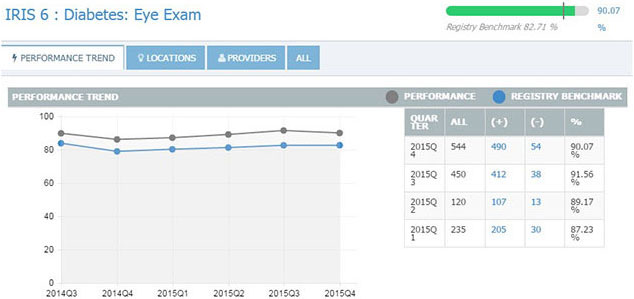
IRIS REGISTRY. Just as individual practices can view their own performance (above, quarterly results for the Diabetes Eye Exam measure), researchers can mine deidentified patient data from the IRIS Registry to conduct studies.
|
Registries: Mining Data
The Academy IRIS Registry (Intelligent Research in Sight), launched in March 2014, is a centralized data repository and reporting tool—the nation’s first electronic health record (EHR)–based comprehensive eye disease and condition registry.
How the IRIS Registry aggregates data. Participating practices integrate their EHR system with the IRIS Registry, which then extracts deidentified data directly from the EHR. “The electronic medical record capture across the heterogeneous software platforms with IRIS is very robust,” said Dr. Adamis. “And the registry’s aim in the future is to collect even more types of information such as imaging data.”
Data and more data. The IRIS Registry makes it easy for practices to monitor their own performance, and it provides researchers with the opportunity to mine mountains of data. “With terabytes of clinical information—derived from nearly 80 million patient visits in just the first 2 years—we could keep PhD students busy nonstop investigating important clinical issues,” said Dr. Parke. “Registry-based research offers us an entirely new avenue to get at some of the questions that previously would have been answered only by RCTs.”
Real-world reflections. Registries like the IRIS Registry can provide excellent data regarding the real-world epidemiology, natural history, and treatment outcomes of various diseases, said Dr. Adamis. “And the larger the N—that is, sample size—the closer you get to the truth.” In addition, Dr. Parke said that access to EHR-derived data helps to evaluate real-world clinical practice, where individuals do things slightly differently, for example, in testing visual acuity (VA). Such variations can have a substantial impact in a small trial, but they become less significant with the large numbers available from registries.
Cheaper and faster. “When the numbers are so large, you have an extraordinarily powerful dataset,” said Dr. Parke, pointing to an example from cardiology: the Thrombus Aspiration in ST-Elevation Myocardial Infarction in Scandinavia (TASTE) trial. Using the national Swedish Coronary Angiography and Angioplasty Registry (SCAAR), the TASTE trial enrolled almost all Scandinavian patients who were about to undergo an unusual coronary intervention.1 “The trial enrolled each patient for under $50 and achieved highly statistically significant results in a tiny fraction of the time a clinical trial or chart review would have taken,” he said.
Patient privacy must be protected. Patient privacy is a principal concern, said Dr. Parke, which is why raw data in the IRIS Registry for use by researchers are stripped of patient identifiers. “And because we have an obligation to our members and their patients, we do not make the database available for another party to do analytics outside our control.”
No substitute for RCTs. Still, “registry-based trials won’t supplant clinical trials with investigational drugs in the near future,” said Dr. Adamis. “It’s difficult to have masked therapies and to monitor data quality, but registries are definitely moving in the right direction.”
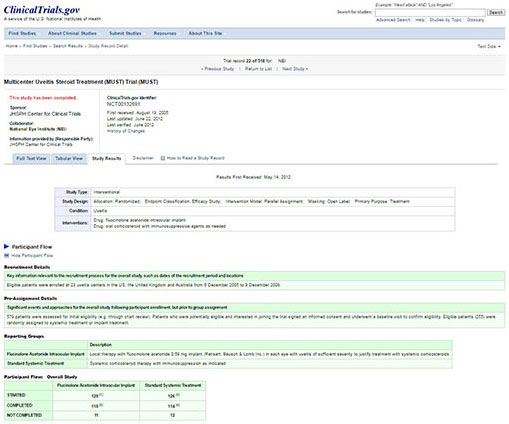 |
CLINICALTRIALS.GOV. Eight years after the FDA made it mandatory in 2007 to report to the ClinicalTrials.gov, rates of compliance are only 13% according to a 2015 New England Journal of Medicine article. 6
|
Data Sharing: Benefits and Formidable Challenges
In 2015, the Institute of Medicine (IOM) issued a report entitled Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risk, which stated that data sharing could advance science by “maximizing the knowledge gained from data collected in trials, stimulating new ideas for research, and avoiding unnecessarily duplicative trials.”2 Because data sharing can refer to a variety of types of data—including study participants’ raw data, metadata, or summary-level data, such as results posted in publications—the IOM noted that future policies will need to stipulate exactly what data will be shared, as well as when and under what conditions.
ICMJE proposal. In early 2016, the International Committee of Medical Journal Editors proposed new requirements for sharing data from clinical trials. These include a definition of data to be released (“the IPD [deidentified individual-patient data] required to reproduce the article’s findings, including necessary metadata”) and the timing of its release (“no later than 6 months after publication”), as well as stipulations regarding how data will be housed and shared. After considering feedback to its proposals (deadline for feedback was April 18), ICMJE will adopt new requirements. These will go into effect a year after adoption to allow investigators time to implement necessary systems.3
Goals of data sharing. “Recognizing that the goal is laudable,” said George B. Bartley, MD, editor-in-chief of Ophthalmology, “our journal encourages data sharing, as indicated in an excerpt from our Guide to Authors: To promote transparency and opportunities for further research, authors of work published in Ophthalmology are encouraged to provide access to relevant datasets in compliance with contemporary reporting standards. …”
In brief, the ICMJE suggests that sharing data will enable independent confirmation of results and increase confidence in conclusions drawn from clinical trials; foster testing of new hypotheses; increase efficiency; and benefit patients, investigators, sponsors, and society at large.3
Build on knowledge. “Everybody likes the idea of data sharing because it advances knowledge,” said Dr. Chambers, “which moves society forward.” Whether results are positive or negative, such sharing is beneficial, said Dr. Chew, adding that negative data can save investigators from going down a fruitless pathway. Unfortunately, less than half of all clinical trials are actually published.4
Data sharing is helpful for industry as well, said Dr. Adamis, using the example of the Genentech drug rituximab, which has treatment of granulomatosis with polyangiitis (formerly Wegener’s granulomatosis) and microscopic polyangiitis among its approved indications. “After reviewing our open-source data online, including patients’ flow cytometry data, Atul Butte, MD, PhD, an independent researcher from University of California, San Francisco, came up with a novel predictive biomarker for our drug that we had never identified.” (See reference 5.)
Improve patient care. Improving patient care is the bottom line for all involved. “Ultimately, sharing data will advance medical knowledge and very likely get more therapies to patients,” said Dr. Adamis. Data sharing should speed innovation and translate into more accurate assessments of therapies’ benefits and risks.
Share the costs. Things have changed since the early days when Dr. Patz borrowed money from his brother-in-law to conduct his studies in patients with ROP, said Dr. Chew. “Clinical research is very expensive, so it makes sense to share our riches [data] and make sure everybody benefits from it. Our most recent study on the genetics of macular degeneration enrolled nearly 34,000 participants worldwide, including cases and controls. We needed that many to really look at rare genetic variants associated with AMD. It takes more than a village—it takes the whole world.”
Challenges. At the top of the list of concerns about or challenges with data sharing is patient privacy. Other considerations include the following.
Lack of standardization. Sharing data is challenging without standardized taxonomy, nomenclature, and definitions, said Dr. Bartley. Different branches of medical research use different terms. Although controlled vocabularies such as MESH and SNOMED are available, they are not applied consistently. How does the medical community reach consensus on basic issues such as this?
Then there is the question of where to deposit different kinds of data—from genomic data and chemical compounds to imaging and algorithm data. “In theory, the idea of data sharing is attractive,” Dr. Bartley said, “but successful execution will be challenging absent a centralized, standardized, easy-to-use repository.”
Onus on researchers. A major hurdle for researchers is a lack of time and resources. “In the real world, those without a lot of resources face huge obstacles when uploading data for even a small trial,” said Dr. Chew. “With our large trials, it has taken the better part of a year to prepare everything.”
Dr. Bartley is concerned that the burden of responsibility falls disproportionately on researchers and authors. Lessons should be learned, he said, from the experiences with ClinicalTrials.gov, launched as a voluntary registry in the 1990s, which the FDA later made mandatory. A recent study published in the New England Journal of Medicine6 found that compliance among researchers was just over 13% between 2009 and 2013—down from 22% cited in an earlier British Medical Journal article.7 “For now, both incentives and penalties are lacking—so registration is basically on the honor system,” he said.
Negative mindsets. Data sharing tends to be something people want others to do, said Dr. Chambers, but they don’t necessarily want to share what they have with other people. Some do “play well in the sandbox,” added Dr. Bartley, but others can be protective of their work and will do the bare minimum to fulfill publication requirements. “Can you blame them?” he asked. “Maybe not.”
In a New England Journal of Medicine editorial supporting data sharing done right, the authors acknowledge that some researchers may have concerns about third-party analysis of their data. Two of these concerns are that 1) “... someone not involved in the generation and collection of the data may not understand the choices made in defining the parameters” and 2) the practice of data sharing may breed a new class of researcher who uses others’ data “for their own ends”—possibly going so far as to use the data to disprove the original researchers’ conclusions.8
Disincentive to gather data? “If you take away the potential benefits [competitive advantage] of having a [proprietary] dataset, will people stop generating those datasets?” asked Dr. Chambers. “Will this decrease incentive for original research?”
Dr. Adamis doesn’t think so. “My sense is that more open data policies will accelerate, not slow, research,” he said. “Data sharing can move forward with the proprietary model that already exists—filing for intellectual property first and then publishing.”
Timing of release. Industry may be concerned about the confidentiality of the proprietary data it submits to the FDA. But Dr. Chambers explained that, as dictated by Congress, the FDA keeps information confidential during the initial phases of the new drug approval process. Only after a product is approved does the FDA publish its data summary on the Web, providing its rationale for approval. “If a product is turned down, none of this information is available,” he said. “Timing of the release of information is important to prevent loss of trade secrets.”
Flawed secondary analyses. Given the complexities of research, it’s too easy to “make mischief” with data—whether intentional or not. Dr. Chambers gave an example from a conference he recently attended, where researchers presented a graph of neonatal outcomes at 100 hospitals. At first blush, the hospital with the lowest mortality looked most favorable, he said. What the data did not explain is that, right across the street from the hospital, there is a tertiary care center, which took all of the high-risk procedures. Thus, the favorable rate was probably influenced by the types of cases treated, not just the quality of the care.
 |
|
NEI FUNDING. Researchers at the NEI may need to become more resourceful in the face of decreases in funding.
|
Future Trends in Research
“My personal view,” said Dr. Parke, “is that we are evolving from the past, when we had basically only 2 weapons: retrospective case series and large multicenter RCTs. Now we have several other options. The future of clinical research isn’t in purely registry-based prospective trials or retrospective data analyses or meta-analyses or RCTs. It will be in finding the best spot for each of these trial structures and in identifying new approaches.”
RCTs still golden. Randomized controlled trials will always be the gold standard for FDA approval of drugs, said Dr. Chew. “I don’t think that passively looking at groups without randomization will pass muster; however, it will be interesting to see if it’s possible to randomize participants for RCTs from a registry.”
But with decreasing funding and shifting priorities at the NIH, she said, researchers will also have to be smarter and more selective, perhaps doing more to leverage other studies—such as deriving diabetic eye disease data from large diabetes trials. For example, networks such as the Diabetic Retinopathy Clinical Research Network (DRCR.net) may be a way to maximize the use of funds from various sources, including government, foundations, and industry, she said. “The DRCR.net is one of the finest examples of how that can work.”
Funneling funding. We now have access to more analytic tools and databases to address clinical questions than we’ve ever had before, said Dr. Parke. “This may help concentrate funding into studies that are most appropriately addressed by RCTs, so it won’t be diluted all the way across the landscape. If the only way to answer a clinical question is through an RCT at a cost of $50,000 per enrolled patient, you will do it. If, on the other hand, it can be better addressed through a registry at $100 a patient, that’s a pretty easy decision, leaving financial resources to apply someplace else.”
Individualization. Research may become more personalized, Dr. Adamis noted. “Now that costs are falling below $1,000, it’s becoming routine to sequence a person’s tumor in oncology. And I can see something similar happening in ophthalmology as well, which potentially means that some therapies will become much more individualized.” Genentech’s phase 3 lampalizumab trials involve assessing patients’ complement genes for polymorphisms and randomizing patients into different cohorts in the study, he said. “However, it may become increasingly difficult to do randomized controlled trials when few patients have the same disease at a molecular level.”
Novel research methods. Researchers are using innovative approaches, for example:
Basket studies and umbrella trials recruit patients based on their molecular status, identifying them through large clinical trial databases, registries, or electronic medical records at major institutions, said Dr. Adamis. These tools could help overcome the problems associated with treating rare mutations.
Basket studies test the effects of a single drug on a single mutation regardless of site of disease origin. In oncology, for example, a basket study may focus on BRAF (B-Raf proto-oncogene serine/threonine kinase gene) in a variety of cancer types (colon, lung, ovarian).
Umbrella studies, on the other hand, test the effects of different targeted drugs on different mutations in a single histological type of cancer.9
“Rather than randomizing, researchers in the future may do small case series with specific mutations to see if treatment outcomes deviate from natural history,” said Dr. Adamis.
Text mining is another novel method, which involves deriving high-quality information from text, said Dr. Chew. There are 2 types of text that are relevant to ophthalmology: biomedical literature and clinical notes. An example of text mining from the literature is GoPubMed, a knowledge-based search engine for biomedical texts.
An example of text mining with clinical notes is the work done using EHRs for genetics discovery by investigators in the eMERGE network.
“Text mining really has 2 roles—extracting information and generating hypotheses,” Dr. Chew said. “Using this technology to extract information from clinical notes has the potential to generate some really exciting tools for clinical decision support. I know from some genetics, Alzheimer’s, and AMD work that text mining biomedical literature brings new clues—perhaps generating new hypotheses for people to explore.”
An explosion of data. Although medical information has been increasing exponentially, it’s going to expand even more dramatically in the future, said Dr. Adamis, especially with the growing availability of wearable biosensors, measuring every imaginable type of physiological output; open-source “-omics” data; and electronic real-world data (e.g., data available through EHRs).
“Getting individual genomes sequenced will create terabytes and petabytes of data,” he said. “We will be buried in data. The future of medical research will be largely defined by those who can best analyze these data. Will we still do hypothesis-driven research, or will we move more to artificial intelligence platforms as they continue to evolve? We are in the middle of a sea change in scientific and medical research, and I think it will have a positive impact on patient care in the future.”
___________________________
1 Fröbert O et al. N Engl J Med. 2013;369(17):1587-1597.
2 Institute of Medicine. Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risk. 2015. Accessed April 25, 2016.
3 Taichman DB et al. JAMA. 2016;315(5):467-468.
4 Chen R et al. BMJ. 2016;352;i637.
5 Nasrallah M et al. Arthritis Res Ther. 2015;17(1):262.
6 Anderson ML et al. N Engl J Med. 2015;372(11):1031-1039.
7 Prayle AP, Smyth AR. BMJ. 2012;344:d7373.
8 Longo DL, Drazen JM. N Engl J Med. 2016;374(3):276.
9 American Association for Cancer Research. Molecularly Informed Clinical Trials. Accessed May 3, 2016.
Data Sharing: On the Right Track?
“I have polled several members of our Ophthalmology Editorial Board about their experiences with data sharing, both from the perspective of depositor and downloader,” said Dr. Bartley, “and the general sense is that ‘we’re not there yet.’ One exception is the PEDIG database, which seems to be relatively successful.”
Pediatric Eye Disease Investigator Group (PEDIG). Formed in 1997 and funded by the NEI, PEDIG is a collaborative network dedicated to facilitating multicenter clinical research in strabismus, amblyopia, and other eye disorders that affect children. More than 100 participating sites with more than 200 pediatric ophthalmologists and pediatric optometrists in the United States, Canada, and the United Kingdom participate in the network.1
At the PEDIG website, you can find a list of the clinical trials and registration numbers and link to information about the studies. “It’s simple and easy to find things [e.g., protocols, summaries of the study, citations], and intuitive to use—a little like looking at a straightforward Excel file,” said Dr. Bartley. “Behind the scenes, people may be scrambling to make data display correctly, but the site’s ease of use has eliminated 1 part of the challenge for the user. Applying that same level of simplicity to other types of data might be a good starting point.”
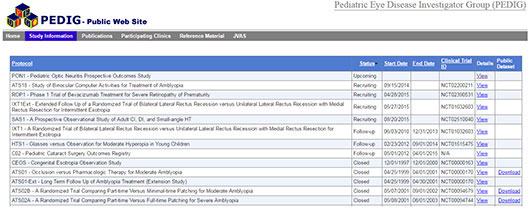
PEDIG. A spreadsheet-style interface makes the PEDIG website easy to navigate.
CrossRef. A nonprofit membership organization that is moving data sharing forward, CrossRef describes its main purpose as promoting the “development and cooperative use of new and innovative technologies to speed and facilitate scholarly research.” It doesn’t hold full text content. Instead, it creates a linking system through which researchers can click on a reference citation in a journal and access the cited article. It seeks to be “the linking backbone for all scholarly information in electronic form.”2
___________________________
1 Pediatric Eye Disease Investigator Group. Accessed April 25, 2016.
2 CrossRef. Accessed April 25, 2016.
|
Meet the Experts
Anthony P. Adamis, MD Senior vice president and global head of Ophthalmology, Immunology, Infectious Disease and Metabolism Clinical Science, Genentech, in South San Francisco, Calif. Relevant financial disclosures: Roche: O.
George B. Bartley, MD Editor-in-chief of Ophthalmology, journal of the American Academy of Ophthalmology, and professor of ophthalmology at the Mayo Clinic in Rochester, Minn. Relevant financial disclosures: None.
Wiley A. Chambers, MD Deputy division director of the Division of Transplant and Ophthalmology Products in the Center for Drug Evaluation and Research at the FDA in Silver Spring, Md. Relevant financial disclosures: None.
Emily Y. Chew, MD Deputy director of the Division of Epidemiology and Clinical Applications at the National Eye Institute, National Institutes of Health in Bethesda, Md. Relevant financial disclosures: None.
David W. Parke II, MD Chief executive officer of the Academy in San Francisco, Calif. Relevant financial disclosures: None.
For full disclosures and the disclosure key, see below.
|